OptiPres II: An AI-Powered Decision Support System with Retrieval-Augmented Generation for Enhancing Optimal Drug Prescription
Posted on Sep 11, 2025 @ 01:33 AM under Python Large Language Models Health Informatics Machine Learning
OptiPres
For my PhD thesis (2015), I tackled a rather interesting and fascinating problem - prescribing optimal personalized drug for patients. The solution I proposed involved developing a mobile intelligent agent architecture that could assist doctors in prescribing the best combination of drugs for a patient in real-time. The main techniques used were an adaptation of the Analytical Hierarchical Process (AHP) and Case-based Reasoning (CBR). Additionally, we needed access to drug data in real-time so the National Drug File Reference Terminology (DFR-RT) API was used for this. To support the implmentation a basic Electronic Medical Record (EMR) system was used. Finally, for the transport layer and embedded intelligence, the Jade-Leap Framework for Mobile Agents was used. This prototype was built using mainly Java with some use of Python for automation and scripting.
This proved to be a difficult task especially since I am not formally trained in medical sciences. As a result, I collaborated with a number of medical professionals and actually got a working prototype - OptiPres.
The details of this work can be seen here.
Below I will provide a summary of the problem and the solution in the following diagrams.
The Problem
From discussions and interviews with a number of health professionals (Doctors, Pharmacists and Nurses), we came up with this concept map that represents the factors that affect prescribing the right drug therapy for patients.
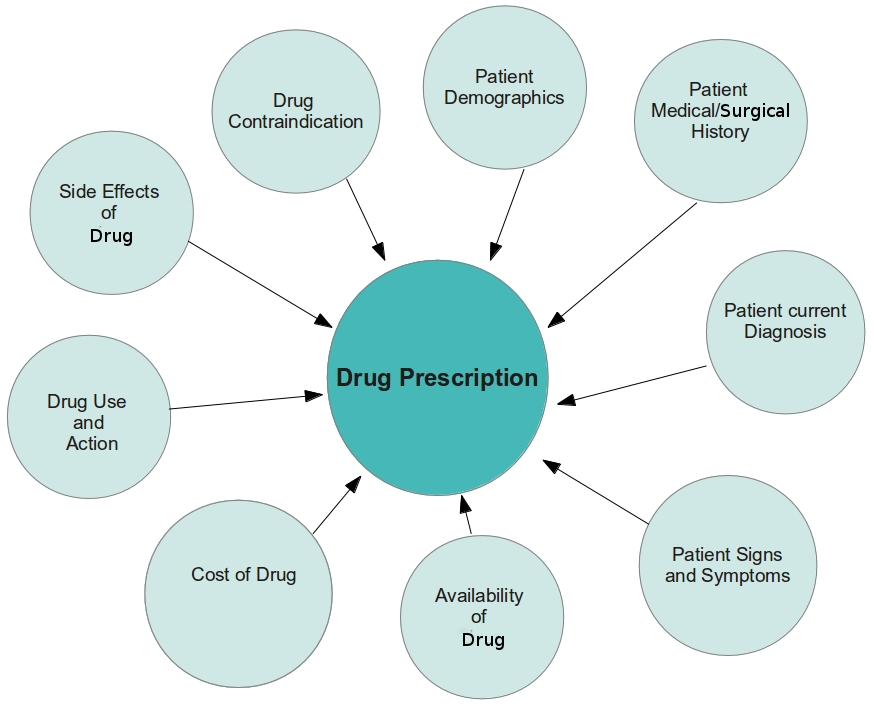
The Solution
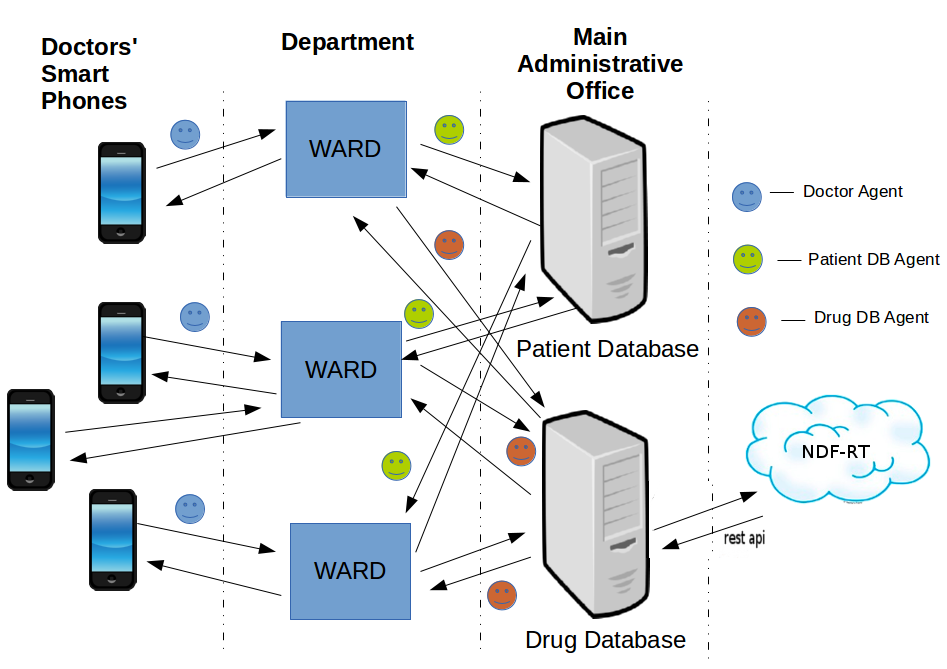
The diagram below respresent a simplified version of the proposed architecture to support the processes to solve this problem.
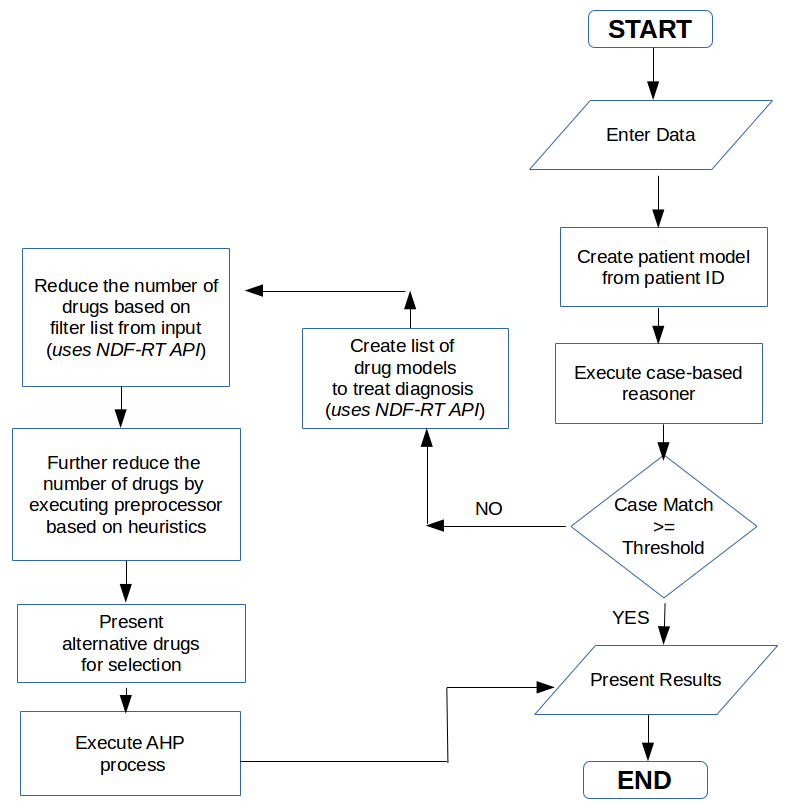
The diagram below shows a flow chart that gives an overview of the steps that were involved in the solution.
OptiPres II
Fast forward to today, I still occasionally come across medical literature highlighting ongoing challenges in effective drug prescription. With the current advancements in AI and available tools, I began to envision how these technologies could significantly enhance OptiPres. This led me to OptiPres II, which uses a more modern approach that uses AI and machine learning techniques. One such technique was the use of Retrieval-Augmented Generation (RAG). RAG basically improves the output of (Large Language Models) LLMs by retrieving and incorporating up-to-date information from external knowledge sources when generating responses.
The aim for this rest of this article is not to present in detail the entire work that is currently being done, but to give the general approach that is being taken in an attempt to assist in solving this problem. This article will not provide the complete code, but will provide some code snippets along the way as this is still early work.
For the machine learning model, the primary focus is on the approach rather than the results, as the current data is synthetic. In the future, this strategy will be validated using real, anonymized datasets. Implementing Retrieval-Augmented Generation (RAG) yielded more accurate results as it enhanced the language model with reliable medical knowledge.
The Solution
A modern approach that uses Machine Learning while augmenting a Large Language Model (LLM) using Retrieval-Augmented Generation (RAG)
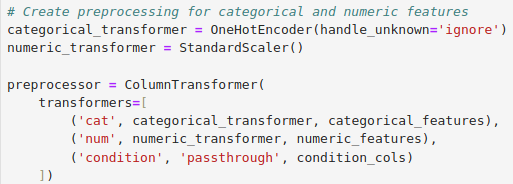
Technologies: - LLM: OpenAI - Jupyter Notebook: Data Analysis - Synthea: Synthetic data base stored in FHIR format - MySQL: Basic EMR to Store Data - Python: Application functionality and Data Analysis - Flask: Web Framework
Creating EMR from FHIR resources using Python
Code snippet uses to process JSON files representing fhir resouces to create tables in database.
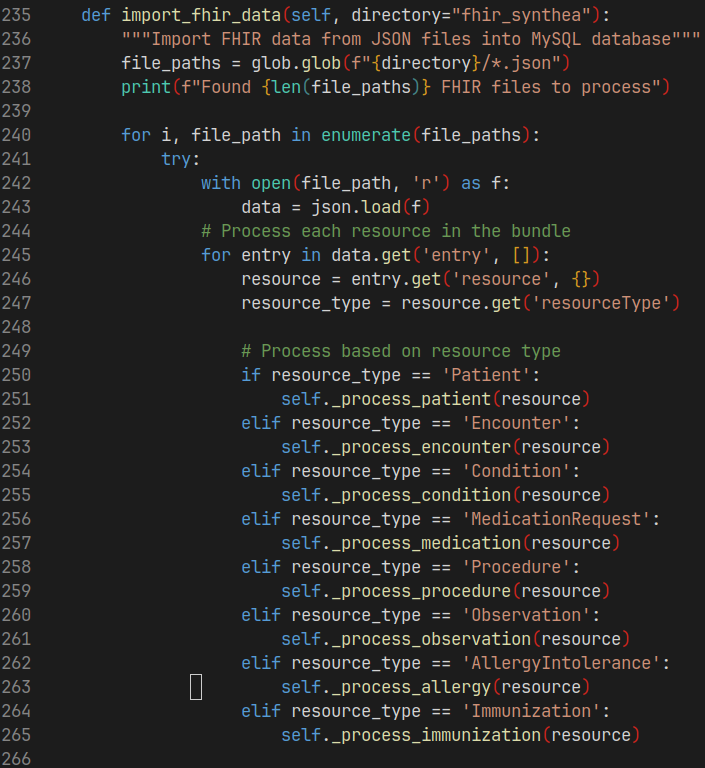
Below shows a snippet of the _process_patient() function:
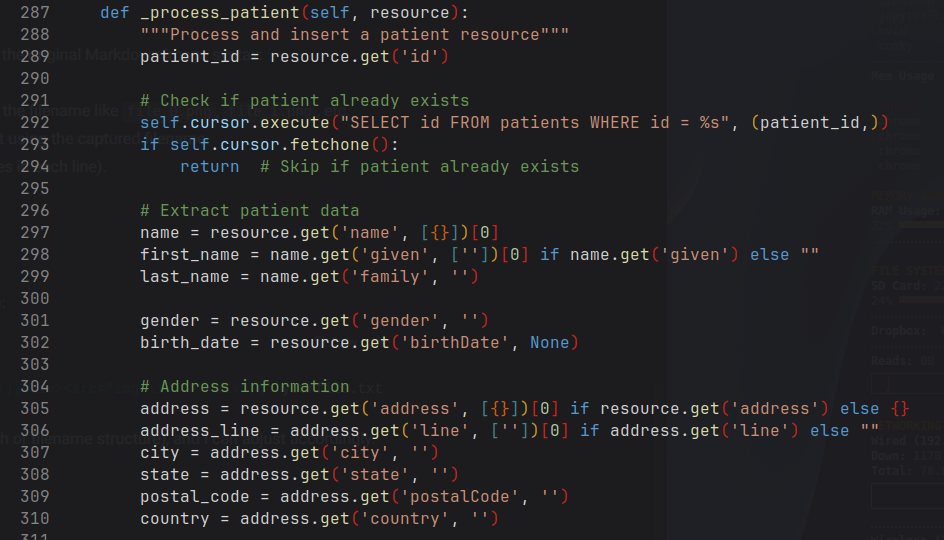
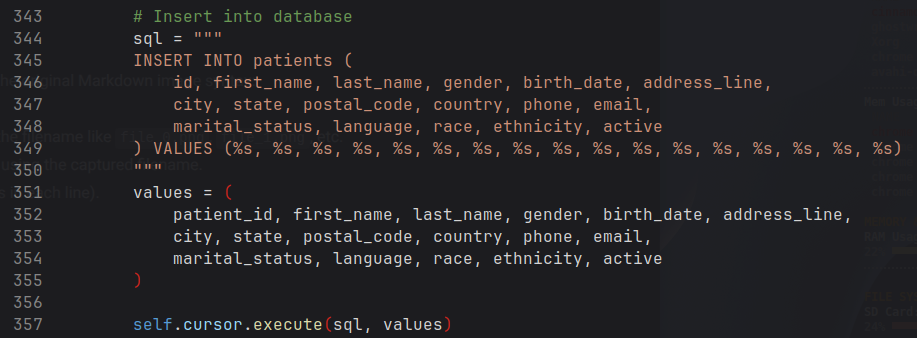
⋮
The created tables are show below:
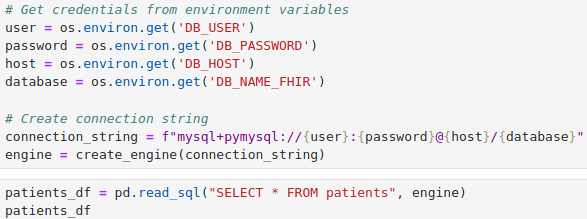
The code from Jupyter Notebook to load data from EMR using the Patient table as as example.
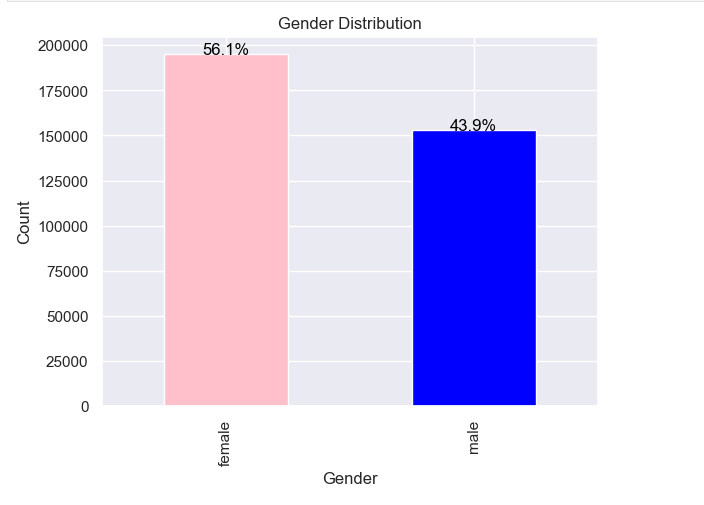
Data Exploration
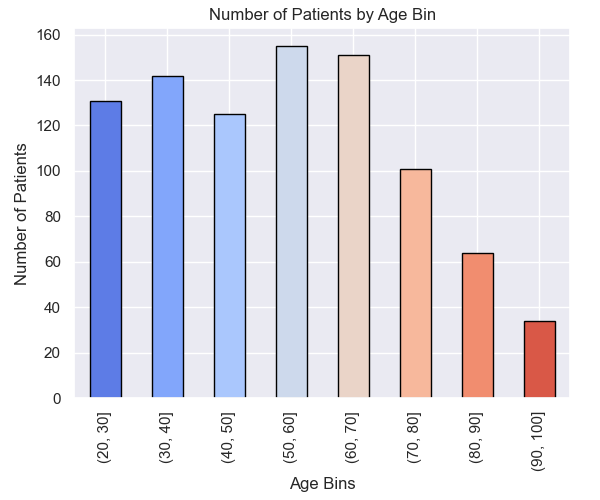
Data Preparation
Feature Engineering



Features

Model Preparation

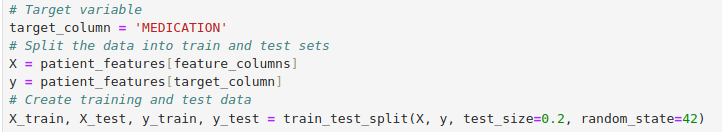
Model
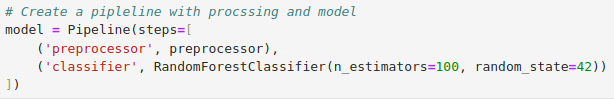

Model accuracy is average 52%, this is not good. Remember that this is synthetic data. However, to improve this, we can try looping through a few other models and perform some hyperparameter tuning using GridSearchCV to see if we can get better results. This is demonstrated below.
Hyperparameter Tuning
Loop through selection of models and select the best model and score

Use GridSearchCV with parameter grid to tune parameters


Save Model
LLM and RAG
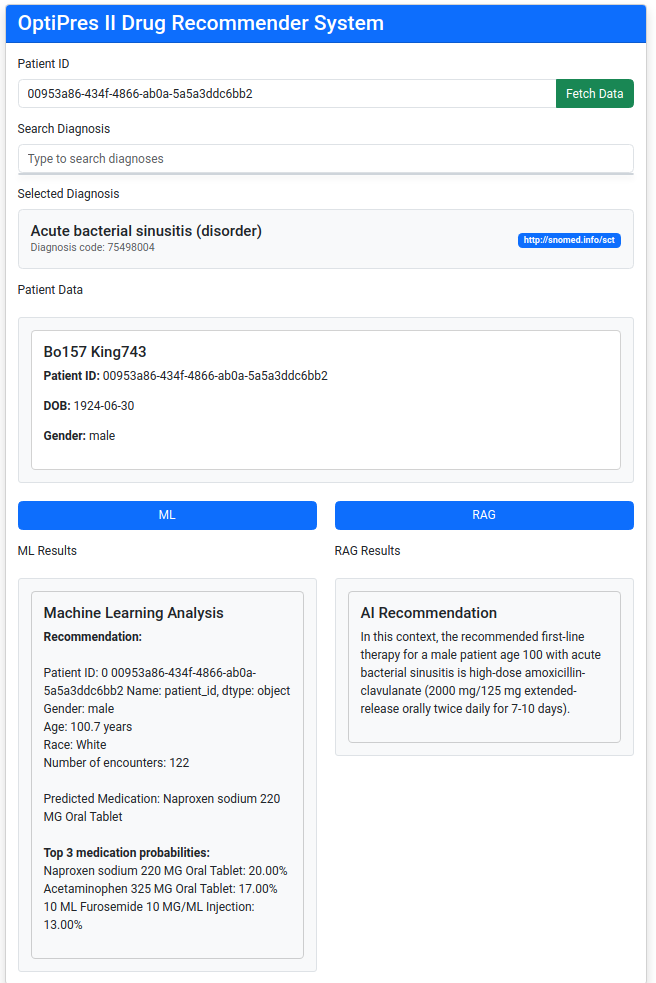
As discussed above, the use of Large Language Models (LLMs) in combination with Retrieval-Augmented Generation (RAG) is expected to yield promising results. Below, we outline the steps taken to implement the RAG solution. For demonstration purposes, a Flask application was developed that supports both strategies, allowing for direct comparison of their outputs.
One key advantage of the RAG approach is that it delivers answers in a conversational format, thanks to the integration of an LLM. Additionally, we plan to include a section that explains the rationale and thought process behind selecting this approach.
Step 1: Create Vector Embeddings from Drug prescription corpus and store in Chroma Database for later querying. - Current Medical Diagnosis and Treatment (SIXTY-FIRST EDITION) - 2022 - Clinical guidelines - Diagnosis and Treatment Manual - 2025 (For curative programmes in hospitals and dispensaries Guidance for prescribing)
Step 2: Get query from user
Step 3: Query vector database and OpenAIEmbeddings to determine sentences that are semantically similar to question asked (This provides context for the LLM).
- OpenAIEmbeddings model convert text into numbers and capture the meaning and context of text)
Step 4: Use prompt template to integrate drug prescription data and patient data with OpenAI ChatGPT API (Local LLM)
Step 5: Send response from Flask server to web application front end.
Considerations for Future Work
Correlation vs. Causation: The model identifies statistical patterns in medication prescribing, not necessarily causal relationships between conditions and appropriate treatments.
Clinical Guidelines: The traditional ML model does not incorporate clinical treatment guidelines or drug interaction information - it is purely based on patterns in the Synthea data.
Multiple Treatment Options: For most conditions, there are multiple appropriate medication options depending on factors like comorbidities, contraindications, and patient-specific factors.
Training Data Limitations: Synthea is synthetic data designed to mimic real-world patterns, but it may not capture the full complexity of clinical decision-making.
Explanation of Results: Many machine learning algorithms function as black boxes, making it challenging to explicitly identify the decision rules or reasoning processes behind the generated solutions.
Security: Ensuring privacy and compliance with regulations like HIPAA.
Decision Support, Not Replacement: This kind of model is better used as a decision support tool rather than an autonomous prescribing system.